Machine learning reveals novel patient subtypes for rare disease
We're using omics to double rare disease patients' chance to respond to therapy.
This is precision medicine.

Machine learning reveals novel patient subtypes for rare disease
Castleman Disease Collaborative Network and Medidata advance potential diagnosis and treatment options with omics data
Summary
For diseases with high unmet need, identifying biomarkers that differentiate patients and determine treatment response can significantly improve outcomes. New machine learning approaches deliver insights into poorly understood patient populations, and these tools are helping researchers identify previously unknown and unconsidered efficacy and safety insights.
Castleman disease patients currently only have one approved therapy with a ~35% response rate. The Castleman Disease Collaborative Network (CDCN) designed the first large serum proteomic study of its kind to:
- Accelerate diagnosis
- Identify which patients will respond to the approved therapy
- Understand etiology of the disease to identify next-generation therapies
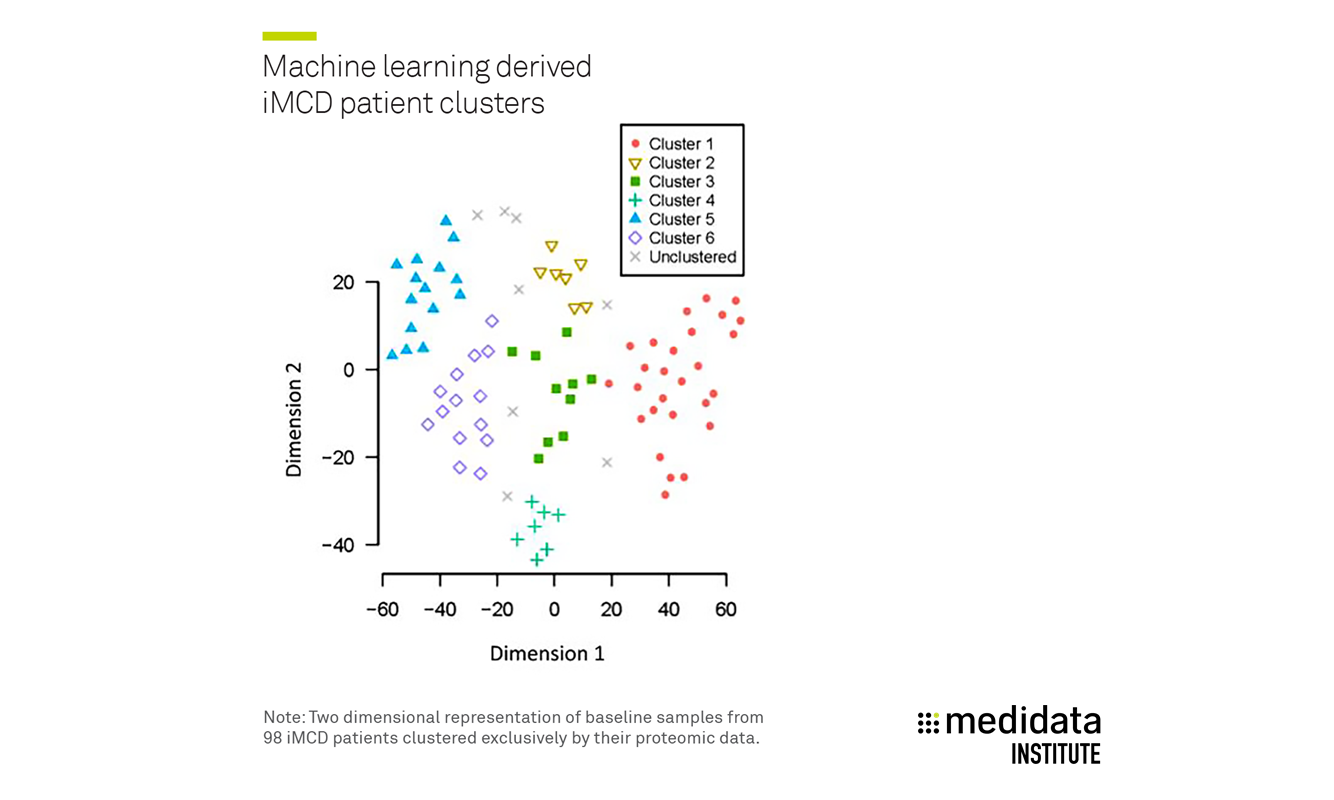
By analyzing baseline proteomic data with machine learning technology, CDCN and Medidata identified six patient clusters previously unknown in idiopathic multicentric Castleman disease (iMCD), including one patient subset that demonstrated a three-time higher response rate than other patients on the approved treatment.
Methodology
CDCN established a 10-party international collaboration of academic groups, non-profits, and industry partners to source blood samples. Samples were collected from iMCD patients, healthy controls, and patients with autoimmune, infectious, and oncological disorders with significant clinical symptomatic and histological overlap to iMCD. SomaLogic performed proteomics assays on all 100+ samples.
Medidata Rave Omics machine learning expertise and technology was used for data integration, quality control, and analysis, including sample clustering of proteomics data.
Key Findings
Medidata and CDCN identified six proteomic clusters of iMCD patients [Figure 1].
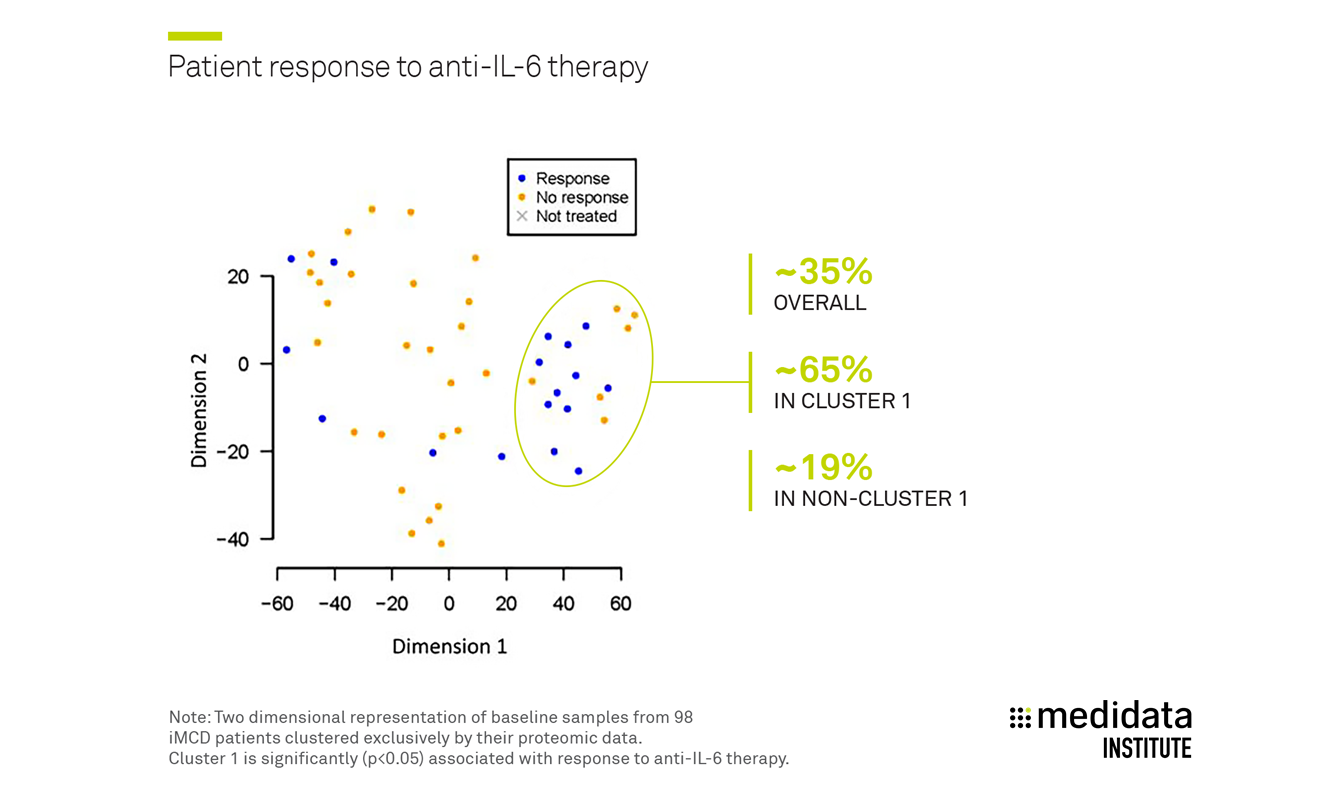
Medidata and CDCN demonstrated that while overall response rate to the existing anti-IL-6 therapy is ~35%, this is primarily driven by one cluster which exhibits a three-fold higher response rate (~65%) compared to other clusters (~19%) [Figure 2].
Implications
Machine learning analysis of omic data can play a significant role in identifying patient subtypes, evidence of biomarkers, and in predicting treatment response in diseases with high unmet need.
Importantly for the rare disease community, the study demonstrates how omic analysis technology can be used to advance research and discover previously unknown treatment options in small populations and subpopulations in larger diseases.
For the iMCD community, the study demonstrated that previously undiscovered proteomically-distinct iMCD groups may exist. CDCN is now working to explore new treatment options based on the proteomic signatures identified in this research.
Learn how the CDCN is accelerating research and treatment for Castleman Disease.